With the recently relased version of the package RGL (V. 0.92.879) there is a new option to publish 3D plots as an interactive WebGL graphic.
Nearly every 3D plot you set up in a RGL window can be exportet via a very easy command. Just plot as usual into your rgl device and then use the command “writeWebGL()”.
Time Space Cube sample:
Dependable on your hardware and software specifications (graphics have to support a newer OpenGL version) you’ll be able to open my try in your browser window:
Click here to open my WebGL sample in a new window (~3MB)
Here the command to export your RGL window to a webpage:
browseURL(paste(“file://”, writeWebGL(dir=file.path(tempdir(), “webGL”), width=700), sep=””))
To get a quick impression about the temporal stay of places it is helpful to generate a plot of the trackpoints spatial density (intensity).
As the 3d visualisation has both advatages and disadvantages, a combination with a 2D plot is useful to interpret the data. The data used in this example is a gps record of the “everyday life” of a test person.
Code snippet:
###################################################
## 3d/2D Plot density with spatstat density.ppp
###################################################
bb_utm <- qbbox(lat = tripdata_utm_num[,2], lon = tripdata_utm_num[,1]) #Boundingbox
Rect <- owin(c(bb_utm$lonR[1]-500, bb_utm$lonR[2]+1800), c(bb_utm$latR[1]-500, bb_utm$latR[2]+500))
P_all <- ppp(tripdata_utm_num[,1], tripdata_utm_num[,2], window=Rect) # opt: marks=datetime
PPP_all <- as.ppp(P_all)
den <- density(PPP_all, sigma = 70)
cutted_den <- den # Wertebereich abschneiden für Extreme
cutvalue <- 0.0020 # Erfahrungswert
cutted_den$v[cutted_den$v>=cutvalue] <-cutvalue
png(paste(plotpath,proband,”intensity_overview_spatstat_v3.png”, sep=””),2400,1200); # Plot Dichte 3D
layout(matrix(c(1,2), 2, 2, byrow = TRUE), widths=c(1,1))
persp(cutted_den , col=”grey”, d=1, ticktype=”detailed”, cex.lab=4, cex.axis=4, zlim=c(range(cutted_den$v)[1],cutvalue), phi=30, theta=-20, xlab=”longitude”, ylab=”latitude”, zlab=”density”, main=paste(proband, “Räumliche Intensität”))
couleurs <- tail(topo.colors(trunc(1.4 * 100)),100)
couleurs[1] <- “#0400ff”
plot(cutted_den , col=couleurs, ticktype=”detailed”, xlab=”longitude”, ylab=”latitude”, cex.lab=5, cex.axis=5, main=paste(proband, “Räumliche Intensität 2D”))
#points(SP_UTM@coords[,1], SP_UTM@coords[,2], cex=0.05, col=”grey”)
points(SP_wp_UTM@coords[,1], SP_wp_UTM@coords[,2], cex=2, col=”red”)
text(SP_wp_UTM@coords[,1], SP_wp_UTM@coords[,2], labels=daten_wp$wpOpen, cex=1, adj=c(0,-1), col=”red”)
dev.off()
# If you use any code, please refer to or cite this site (see about geolabs)
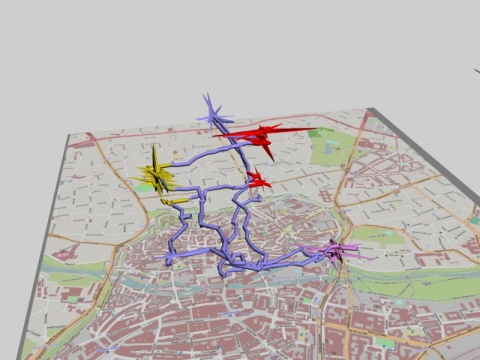
Hägerstrand Time-Space Cube
11/30/2010
With the rgl package it’s possible to interact with the 3d visualization of the timespace tracks.
Code example:
plot3d(lon,lat,timedate, xlim=range(lon), ylim=range(lat), zlim=range(timedate), ticktype=”detailed”, xlab=”longitude”, ylab=”latitude”, zlab=”Date”, col= as.POSIXlt(daten[,”Date”])$mday, type=”l”, main=plottitle)
In the posted example individual waypoints were added by drawing vertical lines.
Many asked for the code. Here is some of it:
################################################# # 2.5 PLOT Hgerstrand TimeSpace 3d interaktiv ################################################# dimen <- dim(daten) dts = daten$Date tms = daten$Time timedate <- timeDate(paste(dts, tms), format = “%Y-%m-%d %H:%M:%S”, zone = “GMT”, FinCenter = “Europe/Berlin”) # timeDate Object erzeugen zshift = 180000 plottitle <- paste(proband, “TimeSpaceCube vom”, range(dts)[1], “bis”, range(dts)[2]) plot3d(lon,lat,as.numeric(timedate+zshift), xlim=range(lon), ylim=range(lat), lit=TRUE, box=FALSE, smooth=TRUE, alpha=c(1), zlim=as.numeric(range(timedate)+zshift), lwd=”3″, axes=T, xlab=””, ylab=””, zlab=””, fog=TRUE, col= as.POSIXlt(daten[,”Date”])$mday%%2+1, type = “l”) # Plot Hgerstrand timespace path / x=longitude , main=plottitle light3d(theta = 0, phi = 15) rgl.bg( sphere = FALSE, fogtype = “none”, color=c(“#FFFFFF”,”white”), back=”lines”) #plot3d(daten_wp$Longitude[6],daten_wp$Latitude[6], timedate[1], size=400, type=”p”) #grid3d(“z+”, at = NULL, col = “#888888″, lwd = 1, lty = 2, n = 8) plot3d(range(lon)[2],lat, timedate, xlim=range(lon), ylim=range(lat), zlim=range(timedate),col=”#CCCCCC”, lwd=”1″, add=T, type=”l”) #sideplot plot3d(lon,lat, range(timedate)[1], xlim=range(lon), ylim=range(lat), zlim=range(timedate),col=”#CCCCCC”, lwd=”1″, add=T, type=”l”) # GroundedPlot #grid3d(“z”, at = NULL, col = “#CCCCCC”, lwd = 1, lty = 2, n = 4) plot3d(daten_wp$Longitude,daten_wp$Latitude, timedate[1]+300, col=”darkgrey”, size=”5″, lwd=”3″, type=”p”, add=T) #plot3d(daten_wp$Longitude,daten_wp$Latitude, range(timedate)[2], col=”darkgrey”, size=”5″, lwd=”3″, type=”p”, add=T) rgl.texts(daten_wp$Longitude,daten_wp$Latitude,range(timedate)[2], color=”black”, text=daten_wp$wpOpen, adj = 0.5, family = “sans”, font = 1, cex = 1, useFreeType=F) daten_wp$id <- c(1:dim(daten_wp)[1]) for(ii in unique(daten_wp[,”id”])){ if (daten_wp$isConstraint[ii] == “TRUE”) { line1 <- c(daten_wp$Longitude[ii], daten_wp$Latitude[ii], as.numeric(timedate[1])[1]) line2 <- c(daten_wp$Longitude[ii], daten_wp$Latitude[ii], as.numeric(range(timedate)[2]+zshift)[1]) lineall <- rbind(line1,line2) lines3d(lineall, lwd=300, col=”black”, alpha=c(0.1)) } } aspect3d(1,1,1.5) # Jeder Proband unterschiedliche Werte zur Anonymisierung snapshot3d(paste(plotpath, proband, “_timespacecube.png”, sep=””)) for (i in 1:60) { count <- seq(1,90,1)[i] rgl.viewpoint(theta=0, phi=count) filename <- paste(plotpath, “\\timespaceani\\pic”,formatC(i,digits=1,flag=”0″),”.png”,sep=””) rgl.snapshot(filename) }
Time-Space Cloud with R
02/22/2010
Here comes another option to analyze a TimeSpace-Track with R. A lattice cloud plots every recorded trackpoint into a 3d-time-space-cube. As the data (planar point pattern) is marked with the daytime, cluster of everyday routines become visible.
Here the direct comparison between a function of density and the time-space-cloud.
Code example:
cloud(time_hours ~ PPP_selection$x * PPP_selection$y, data = daten, zlim = c(23,0), xlim = c(653000,643000), screen = list(z = 160, x = 120), panel.aspect = 0.75, xlab = “Longitude”, ylab = “Latitude”, zlab = “Time”, scales = list(z = list(arrows = FALSE, distance = 2), x = list(arrows =FALSE, distance = 2), y = list(arrows = FALSE, distande = 2)),)
This examle is inspired by: http://lmdvr.r-forge.r-project.org/ (Figure 6.2)
Time-Space Analysis with R
02/06/2010
Beside the visualisation of TimeSpace Tracks, I’m trying to find a way to analyze GPX-Tracks with statistical software. This are the first results with R (The R Project for Statistical Computing):
^This graph is a result of the analysis with the package trip (Spatial analysis of animal track data). Unfortunatelly i’m do not understand witch scale is used by the package.
^Trackpoints as a function of density.
Since there is a trackpoint recorded every 10 sec., it is possible to interpretate the density of the trackpoints as time-spend.
This is a two day track. The highest peak in the right corner is my home (Nuremberg). The peaks in the backstage are both university in Erlangen. The path on the rigth side I did with my bicycle, the left one with the train.
But how to examine specific areas?
^1500 m arround my house in the city center.
With clickppp() from the spatstat package it’s possible to choose e.g. a point with the mouse:
####### Example Code:
plot(tripdata_utm) # plots the recorded trackpoints (converted to UTM)
P_center <- clickppp(n=1, win=Rect, add=TRUE, main=NULL, hook=NULL) # Select a point in the plot with the mouse
center <- as.data.frame(P_center)
D <- disc(radius = 1500, centre = c(center[,1], center[,2])) # create a disc window
P_selection <- ppp(tripdata_utm_num[,1], tripdata_utm_num[,2], window=D) # reduce the data with the window
^Another function of density (2D).
^Trackpoints as a function of time.
Here the trackpoints are divided by a grid and counted. Since the device records the position every 10 sec. The qqcount can be clearly interpreted as time-spend.
The next step is to add this data to a gis layer.